... Amino-terminal processing in the yeast Saccharomyces cerevisiae has been investigated by examining numerous mutationally altered forms of iso-1-cytochrome c. Amino-terminal residues of methionine were retained in sequences having penultimate residues of arginine, asparagine, glutamine, isoleucine, leucine, lysine, and methionine; in contrast, the amino-terminal methionine residues were exercised from residues of alanine, glycine, and threonine and were partially excised from residues of valine. ...
This conclusion is consistent, in general, with the deduced specificity of the enzyme based on the analysis of known amino-terminal sequences of intracellular proteins (S. Tsunasawa, J. W. Stewart, and F. Sherman, J. Biol. Chem. 260:5382-5391, 1985).
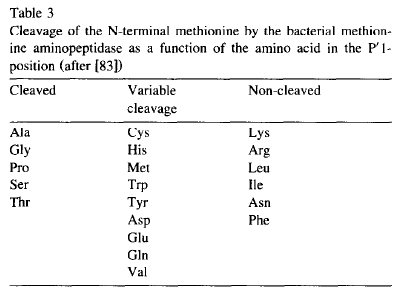
Gonzales and Robert-Baudouy showed a table in their review paper (FEMS Microbiol Rev. 1996 Jul;18(4):319-44) about MAP.
One good way to ease these criticisms is to make bioethanol from cellulosic biomass. Last month, DOE (the Department of Energy) announced that they backed 385 milion dollars to six companies making cellulosic bioethanol.
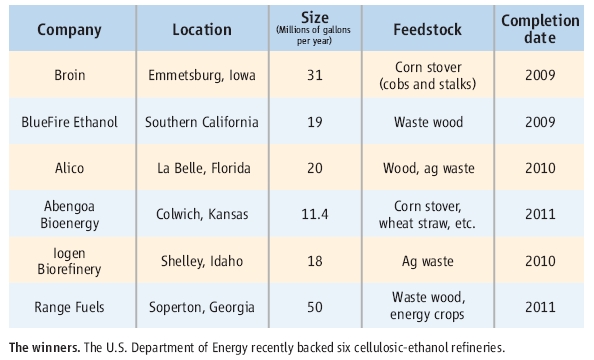
The country of bioethanol has been Brazil which produces 4.2 billion gallons a year from sugar cane. US can produce almost 4 billion gallons of bioethanol from corn. However, the six companies can only produce 0.13 billion gallons of cellosic bioethanol. It is still very small portion of total bioethanol production but if it works good, it could be very strong altanative way to produce bioethanol from wasted biomass.
Corn: The inflation crop (CNN Money)
The great corn gold rush (Fortune)
Cellulosic Ethanol:Biofuel Researchers Prepare to Reap a New Harvest (Science)
How Biofuels Could Starve the Poor (Foreign Affairs)
Corn ethanol could hurt poor's food security; (biopact.com)
Basically plasmids with same replication origin are thought to be incompatible. The most commonly used replication origins of E. coli vectors are pBR322-derived ColE1 and P15A replicons. Most of pET expression vectors contain ColE1 origin and pACYC derived plasmids contain P15A origin. However, several other origins can be used for multi plasmid experiments.
Recently, Novagen which developed the pET expression system launched DUET vectors. Each DUET vector has double T7 promoters and double multicloning sites. There are five vectors of DUET vectors with four diffferent origins.
- pACYCDuet-1 carries the P15A replicon, lacI gene and chloramphenicol resistance gene.
- pCDFDuet-1 carries the CloDF13 replicon, lacI gene and streptomycin/ spectinomycin resistance gene (aadA).
- pCOLADuet-1 carries the ColA replicon, lacI gene and kanamycin resistance gene.
- pETDuet-1 carries the pBR322-derived ColE1 replicon, lacI gene and ampicillin resistance gene.
- pRSFDuet-1 carries the RSF1030 replicon, lacI gene and kanamycin resistance gene.
The most famous but old review paper about plasmid incompatibility is "Plasmid Incompatibility, by RICHARD P. NOVICK (MICROBIOLOGICAL REVIEWS, Dec. 1987, p. 381-395)
But recently an interesting paper came out. The title is "Plasmid incompatibility: more compatible than previously thought?" The authors re-examined plasmid incompatibility concept in terms of the plasmid copy number, by introducing plasmids containing the same origin of replication and different antibiotic resistance genes into bacteria. And they showed that higher copy number leads to longer persistence, but even with low copy plasmids, persistence occurs to a significant degree.
In most cases, unless the gene is correct, your proteins might be modified after translation. Post-translational modification is pretty common in any organisms, especially in eukayotes. Even if you express your protein in E. coli, you may get somewhat different MASS results.
The web site below is very useful to identify what kind of modification was occurred in your protein from molecular weight difference.
http://www.abrf.org/index.cfm/dm.home
If you want to see all kinds of modifications,
Which Superhero are you?
Click above link and answer several questions, then you can find your potential.
Do you want to know who I am?
If you click the links you can get more detailed information from the companies.
1) PreScission Protease (GE Health Care)
PreScission Protease is a genetically engineered fusion protein consisting of human rhinovirus 3C protease and GST (1). This protease was specifically designed to facilitate removal of the protease by allowing simultaneous protease immobilization and cleavage of GST fusion proteins produced from the pGEX-6P vectors pGEX-6P-1, pGEX-6P-2, and pGEX-6P-3; see pGEX Vectors (GST Gene Fusion System). PreScission Protease specifically cleaves between the Gln and Gly residues of the recognition sequence of LeuGluValLeuPheGln/GlyPro (2).
2) TEV (Invitrogen)
AcTEV™ Protease is an enhanced form of Tobacco Etch Virus (TEV) protease that is highly site-specific, highly active, and significantly more stable than native TEV protease, resulting in long-term activity. AcTEV™ Protease specifically recognizes a seven amino acid (Glu-Asn-Leu-Tyr-Phe-Gln-Gly, cleaving between Gln and Gly), making it useful for removing affinity tags from fusion proteins (1,2). For proper cleavage, the protein of interest is expressed with an AcTEV™ (TEV) Protease recognition sequence located between it and the affinity tag. Subsequent incubation with AcTEV™ Protease releases the protein of interest from the fusion tag. This is an effective way to remove solubility, secretion, detection, and purification tags from recombinant proteins. AcTEV™ Protease features
Enhanced enzyme stability for prolonged protease activity
Activity over a broad temperature range (+4°C to 30°C)
Activity over a broad pH range (pH 6.0 to 8.5)
A six-histidine sequence to facilitate its removal from the digested protein sample
Greater than 85% single-band purity with no non-specific protease contamination
3) Factor Xa (Qiagen, Novagen, Sigma etc)
Restriction Grade Factor Xa is a highly purified enzyme isolated from bovine plasma and activated with Russell’s viper venom. The Novagen preparation is purified to near homogeneity and shows no secondary cleavage from contaminating proteases. The preparation is also functionally tested for activity with fusion proteins.
Like enterokinase, Factor Xa cleaves at the C-terminal side of its recognition sequence (IleGluGlyArg↓) and can, therefore, be used for removing all vector-encoded sequences from appropriately designed constructs.
4) Thrombin (Novagen, Sigma, etc)
Restriction Grade Thrombin is qualified to specifically cleave target proteins containing the recognition sequence LeuValProArg↓GlySer. The preparation is functionally tested for activity with fusion proteins and is free of detectable contaminating proteases. Thrombin is supplied with 10X Thrombin Cleavage Buffer and a Cleavage Control Protein.
5) SUMO Protease (Invitrogen)
SUMO Protease, also known as Ulp, is a recombinant fragment of ULP1 (Ubl-specific protease 1) from Saccharomyces cerevisiae. It is highly specific for the SUMO protein fusion, recognizing the tertiary structure of SUMO rather than an amino acid sequence.
6) HRV 3C Protease (Novagen)
Recombinant type 14 3C protease from human rhinovirus (HRV 3C) is the newest addition to the Novagen brand line of restriction grade proteases. The recombinant protease is a highly purified recombinant 6XHis-fusion protein, which recognizes the same cleavage site as the native enzyme: LeuGluValLeuPheGln/GlyPro. The small, 22 KDa size of the protease, optimal activity at 4° C, high specificity, and His•Tag® fusion make HRV 3C protease an ideal choice for rapid removal of fusion tags. The pET expression vectors pET-47b(+) through pET-50b(+) incorporate the HRV 3C protease cleavage site in combinations with His•Tag, S•Tag, thioredoxin (Trx•Tag™), glutathione-S-transferase (GST•Tag™), and NusA (Nus•Tag™) sequences. The combination of pET-47b(+) to pET-50b(+) vectors for expression, HRV 3C protease for fusion tag cleavage, and Ni-NTA His•Bind® metal affinity chromatography for protein purification as well as protease and fusion tag removal allows production of recombinant proteins free of vector-encoded sequences. Supplied as: 2000 units/ml in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 0.5 mM THP, and 50% glycerol.
7) TagZyme, DAPase (Qiagen)
The TAGZyme System removes N-terminal His tags from recombinant proteins with high specificityand efficiency. DAPase Enzyme is used to sequentially cleave off dipeptides from the N-terminus of a purified, His-tagged protein (see figure "His-tag Removal Using TAGZyme Enzymes A", steps 1 and 2). Digestion is halted when the enzyme reaches a “stop point”, an amino acid motif that cannot serve as a substrate (see table "DAPase stop points").
8) Enterokinase (Novagen)
Recombinant Enterokinase (rEK) is a highly purified preparation of the catalytic subunit of recombinant bovine enterokinase, which recognizes the identical cleavage site as the native enzyme (i.e., AspAspAspAspLys↓) and has similar enzymatic activity. rEK exhibits superior rates of cleavage of fusion proteins containing the recognition sequence when compared to the native enzyme (Collins-Racie 1995). Novagen rEK is purified to near homogeneity and, unlike some preparations of native bovine enterokinase, exhibits no secondary cleavage arising from contaminating proteases. The preparation is also functionally tested for activity with fusion proteins.




